Tutorial for PyDrugLogics#
This tutorial demonstrates the use of PyDruglogics for optimizing of Boolean Models and predict drug synergy effects. The PyDruglogics pipeline implementation on based on: Å. Flobak, J. Zobolas (2023): Fine tuning a logical model of cancer cells to predict drug synergies: combining manual curation and automated parameterization.
More information about the package on GitHub.
Core Features#
Construct Boolean model from
.siffileLoad Boolean model from
.bnetfileOptimize Boolean model
Generate perturbed models
Evaluate drug synergies
Initialization, loading files#
The parameters and the initialization of the pipeline are summarized in this section. These are the neccesary components for constructing, training, and applying predictive models effectively.
import pydruglogics
from pydruglogics.model.BooleanModel import BooleanModel
from pydruglogics.model.InteractionModel import InteractionModel
from pydruglogics.input.ModelOutputs import ModelOutputs
from pydruglogics.input.TrainingData import TrainingData
from pydruglogics.input.Perturbations import Perturbation
Model Outputs#
Defines the network nodes and their respective weights used to calculate a global output response for the Boolean Model.
Initialization:
From a File: Load target states from a file that specifies node-state pairs.
From a Dictionary: Define directly using a dictionary where each key-value pair represents a node and its integer weight.
model_outputs_dict = {
"RSK_f": 1.0,
"MYC": 1.0,
"TCF7_f": 1.0,
"CASP8": -1.0,
"CASP9": -1.0,
"FOXO_f": -1.0
}
model_outputs = ModelOutputs(input_dictionary=model_outputs_dict)
2024-12-05 23:41:39,560 - INFO - Model outputs are initialized from dictionary.
model_outputs.print()
Model output: RSK_f, weight: 1.0
Model output: MYC, weight: 1.0
Model output: TCF7_f, weight: 1.0
Model output: CASP8, weight: -1.0
Model output: CASP9, weight: -1.0
Model output: FOXO_f, weight: -1.0
Training Data#
Inclues condition-response pairs (observations) and a weight number used to calculate the weighted average fitness of the Boolean Models.
Initialization:
From a File: Load target states from a structured file.
training_data_file = 'training'
training_data = TrainingData(input_file=training_data_file)
2024-12-05 23:41:39,585 - INFO - Training data loaded from file: training.
From a Dictionary: Define directly using a dictionary.
observations = [(["CASP3:0", "CASP8:0","CASP9:0","FOXO_f:0","RSK_f:1","CCND1:1",
"MYC:1","RAC_f:1","JNK_f:0","MAPK14:0","AKT_f:1","MMP_f:1",
"PTEN:0","ERK_f:1","KRAS:1","PIK3CA:1","S6K_f:1","GSK3_f:0",
"TP53:0","BAX:0","BCL2:1","CTNNB1:1","TCF7_f:1","NFKB_f:1"], 1.0)]
training_data = TrainingData(observations=observations)
2024-12-05 23:41:39,606 - INFO - Training data initialized from list.
training_data.print()
Observation:
Condition: -
Response: CASP3:0, CASP8:0, CASP9:0, FOXO_f:0, RSK_f:1, CCND1:1, MYC:1, RAC_f:1, JNK_f:0, MAPK14:0, AKT_f:1, MMP_f:1, PTEN:0, ERK_f:1, KRAS:1, PIK3CA:1, S6K_f:1, GSK3_f:0, TP53:0, BAX:0, BCL2:1, CTNNB1:1, TCF7_f:1, NFKB_f:1
Weight: 1.0
Perturbations#
Defines the drugs and the perturbations applied to the Boolean Models, affecting the nodes in the network.
Initialization From Dictionary:
Define a
drug_dataandperturbation_data: Provide a list of drugs with their name, targets, and effect (Options:inhibits(default) oractivates). Specify a list of perturbations, where each entry is a drug combination.Define only
drug_data: If noperturbation_datais given, perturbations will be generated automatically by creating all possible single-drug and two-drug combinations from the drug panel.
drug_data = [
['PI', 'PIK3CA', 'inhibits'],
['PD', 'MEK_f'],
['CT','GSK3_f'],
['BI', 'MAPK14'],
['PK', 'CTNNB1'],
['AK', 'AKT_f'],
['5Z', 'MAP3K7']
]
Note: The perturbation_data is optional. If only the drug_data is given, all perturbations are calculated.
perturbation_data = [
['PI'],
['PD'],
['CT'],
['BI'],
['PK'],
['AK'],
['5Z'],
['PI', 'PD'],
['PI', 'CT'],
['PI', 'BI'],
['PI', 'PK'],
['PI', 'AK'],
['PI', '5Z'],
['PD', 'CT'],
['PD', 'BI'],
['PD', 'PK'],
['PD', 'AK'],
['PD', '5Z'],
['CT', 'BI'],
['CT', 'PK'],
['CT', 'AK'],
['CT', '5Z'],
['BI', 'PK'],
['BI', 'AK'],
['BI', '5Z'],
['PK', 'AK'],
['PK', '5Z'],
['AK', '5Z']]
perturbations = Perturbation(drug_data=drug_data, perturbation_data=perturbation_data)
2024-12-05 23:41:39,655 - INFO - Drug panel data initialized from list.
2024-12-05 23:41:39,655 - INFO - Drug perturbations initialized.
perturbations.print()
[PI (targets: PIK3CA)]
[PD (targets: MEK_f)]
[CT (targets: GSK3_f)]
[BI (targets: MAPK14)]
[PK (targets: CTNNB1)]
[AK (targets: AKT_f)]
[5Z (targets: MAP3K7)]
[PI (targets: PIK3CA), PD (targets: MEK_f)]
[PI (targets: PIK3CA), CT (targets: GSK3_f)]
[PI (targets: PIK3CA), BI (targets: MAPK14)]
[PI (targets: PIK3CA), PK (targets: CTNNB1)]
[PI (targets: PIK3CA), AK (targets: AKT_f)]
[PI (targets: PIK3CA), 5Z (targets: MAP3K7)]
[PD (targets: MEK_f), CT (targets: GSK3_f)]
[PD (targets: MEK_f), BI (targets: MAPK14)]
[PD (targets: MEK_f), PK (targets: CTNNB1)]
[PD (targets: MEK_f), AK (targets: AKT_f)]
[PD (targets: MEK_f), 5Z (targets: MAP3K7)]
[CT (targets: GSK3_f), BI (targets: MAPK14)]
[CT (targets: GSK3_f), PK (targets: CTNNB1)]
[CT (targets: GSK3_f), AK (targets: AKT_f)]
[CT (targets: GSK3_f), 5Z (targets: MAP3K7)]
[BI (targets: MAPK14), PK (targets: CTNNB1)]
[BI (targets: MAPK14), AK (targets: AKT_f)]
[BI (targets: MAPK14), 5Z (targets: MAP3K7)]
[PK (targets: CTNNB1), AK (targets: AKT_f)]
[PK (targets: CTNNB1), 5Z (targets: MAP3K7)]
[AK (targets: AKT_f), 5Z (targets: MAP3K7)]
Initialize Boolean Model#
A BooleanModel defines the network and logical rules for node interactions, used for generating predictions and simulations.
There are two ways for the initialization:
Initialize Boolean Model from .sif file#
Creating an InteractionModel, that can be initialized:#
interaction_file(required): The path to the.siffile.model_name: name of the model, by default the.siffile name will be set.remove_self_regulated_interactions: Remove self regulated interactions. The default isFalse.remove_inputs: Trim inputs from the model. The deafult isFalse.remove_outputs: Trim ouputs from the model. The default isFalse.
Creating a BooleanModel, that can be initialized:#
model(required): Interaction Modelmodel_name: The name of the model, by default the.siffile name will be set.mutation_type: There are 3 options:topology,mixed,balanced. The default isbalanced.attractor_tool: There are 2 options:mpbnorpyboolnetThe default ismpbn. More infotmation here about MPBN (Most Permissive Boolean Networks) and more information here about PyBoolNet.attractor_type: There are 2 options:stable_statesortrapspaces. The default isstable_states.
network_sif = 'network.sif'
model = InteractionModel(interactions_file=network_sif)
model.print()
2024-12-05 23:41:39,678 - INFO - Interactions loaded successfully
Target: Antisurvival, activating regulators: FOXO_f, CASP3
Target: CASP3, activating regulators: CASP8, CASP9
Target: FOXO_f, inhibitory regulators: NLK, AKT_f
Target: Prosurvival, activating regulators: CCND1, MYC
Target: CCND1, activating regulators: TCF7_f, RSK_f
Target: MYC, activating regulators: TCF7_f
Target: RAC_f, activating regulators: mTORC2_c, DVL_f
Target: mTORC2_c, activating regulators: TSC_f, inhibitory regulators: S6K_f
Target: DVL_f, activating regulators: FZD_f
Target: MAP3K4, activating regulators: RAC_f
Target: MAP3K11, activating regulators: RAC_f
Target: MAP2K4, activating regulators: MAP3K4, MAP3K11, GRAP2, MAP3K7
Target: MAP3K7, activating regulators: TAB_f
Target: GRAP2, inhibitory regulators: MAPK14
Target: MAP2K7, activating regulators: GRAP2, MAP3K7
Target: JNK_f, activating regulators: MAP2K7, MAP2K4, inhibitory regulators: DUSP1
Target: DUSP1, activating regulators: MSK_f, MAPK14
Target: MAPK14, activating regulators: MAP2K3, MAP2K4, inhibitory regulators: DUSP1
Target: MAP2K3, activating regulators: MAP3K5, MAP3K7
Target: MAP3K5, inhibitory regulators: AKT_f
Target: AKT_f, activating regulators: mTORC2_c, PDPK1
Target: TAB_f, inhibitory regulators: MAPK14
Target: MSK_f, activating regulators: ERK_f, MAPK14
Target: RTPK_f, activating regulators: MMP_f, RTPK_g, inhibitory regulators: MEK_f, MAPK14
Target: RTPK_g, activating regulators: FOXO_f
Target: MEK_f, activating regulators: RAF_f, MAP3K8, inhibitory regulators: ERK_f
Target: SHC1, activating regulators: RTPK_f, inhibitory regulators: PTEN
Target: PTEN, activating regulators: PTEN_g, inhibitory regulators: GSK3_f
Target: GRB2, activating regulators: SHC1
Target: SOS1, activating regulators: GRB2, inhibitory regulators: ERK_f
Target: ERK_f, activating regulators: MEK_f, inhibitory regulators: DUSP6
Target: KRAS, activating regulators: PTPN11, SOS1
Target: PTPN11, activating regulators: GAB_f
Target: RAF_f, activating regulators: KRAS, inhibitory regulators: RHEB, ERK_f, AKT_f
Target: RHEB, inhibitory regulators: TSC_f
Target: MAP3K8, activating regulators: IKBKB
Target: DUSP6, activating regulators: ERK_f, mTORC1_c
Target: mTORC1_c, activating regulators: RHEB, RSK_f, inhibitory regulators: AKT1S1
Target: GAB_f, activating regulators: GRB2, inhibitory regulators: ERK_f
Target: RSK_f, activating regulators: ERK_f, PDPK1
Target: PDPK1, activating regulators: PIK3CA, inhibitory regulators: PTEN
Target: PIK3CA, activating regulators: KRAS, GAB_f, IRS1
Target: IRS1, inhibitory regulators: IKBKB, S6K_f, ERK_f
Target: S6K_f, activating regulators: PDPK1, mTORC1_c
Target: IKBKB, activating regulators: MAP3K7, inhibitory regulators: TP53
Target: PTEN_g, activating regulators: EGR1
Target: GSK3_f, inhibitory regulators: ERK_f, AKT_f, MAPK14, DVL_f, LRP_f, S6K_f, RSK_f
Target: TSC_f, activating regulators: GSK3_f, inhibitory regulators: IKBKB, ERK_f, AKT_f, RSK_f
Target: TP53, activating regulators: MAPK14, inhibitory regulators: MDM2
Target: AKT1S1, inhibitory regulators: AKT_f
Target: MDM2, activating regulators: MDM2_g, AKT_f, inhibitory regulators: S6K_f
Target: MDM2_g, activating regulators: TP53, NFKB_f
Target: BAX, activating regulators: TP53
Target: CYCS, activating regulators: BAX, inhibitory regulators: BCL2
Target: BCL2, inhibitory regulators: BAD
Target: BAD, inhibitory regulators: AKT_f, RSK_f
Target: NLK, activating regulators: MAP3K7
Target: CASP9, activating regulators: CYCS
Target: CASP8, inhibitory regulators: CFLAR
Target: CFLAR, activating regulators: AKT_f, inhibitory regulators: ITCH
Target: ITCH, activating regulators: JNK_f
Target: FZD_f, inhibitory regulators: SFRP1
Target: SFRP1, activating regulators: SFRP1_g
Target: LRP_f, activating regulators: FZD_f, JNK_f, ERK_f, MAPK14, inhibitory regulators: DKK_f
Target: DKK_f, activating regulators: DKK_g
Target: AXIN1, inhibitory regulators: LRP_f
Target: CK1_f, inhibitory regulators: LRP_f
Target: BTRC, activating regulators: GSK3_f, CK1_f, AXIN1
Target: CTNNB1, activating regulators: CHUK, inhibitory regulators: BTRC
Target: CHUK, activating regulators: AKT_f
Target: TCF7_f, activating regulators: CTNNB1, inhibitory regulators: NLK
Target: EGR1, inhibitory regulators: TCF7_f
Target: DKK_g, activating regulators: TCF7_f, inhibitory regulators: MYC
Target: SFRP1_g, inhibitory regulators: MYC
Target: NFKB_f, activating regulators: IKBKB, CHUK, MSK_f
Target: LEF, activating regulators: CTNNB1
Target: MMP_f, activating regulators: LEF
boolean_model_sif = BooleanModel(model=model, model_name='test1',
mutation_type='balanced', attractor_tool='mpbn', attractor_type='stable_states')
2024-12-05 23:41:39,693 - INFO - Boolean Model from Interaction Model is created.
boolean_model_sif.print()
Antisurvival *= (FOXO_f or CASP3)
CASP3 *= (CASP8 or CASP9)
FOXO_f *= not (NLK or AKT_f)
Prosurvival *= (CCND1 or MYC)
CCND1 *= (TCF7_f or RSK_f)
MYC *= (TCF7_f)
RAC_f *= (mTORC2_c or DVL_f)
mTORC2_c *= (TSC_f) and not (S6K_f)
DVL_f *= (FZD_f)
MAP3K4 *= (RAC_f)
MAP3K11 *= (RAC_f)
MAP2K4 *= (MAP3K4 or MAP3K11 or GRAP2 or MAP3K7)
MAP3K7 *= (TAB_f)
GRAP2 *= not (MAPK14)
MAP2K7 *= (GRAP2 or MAP3K7)
JNK_f *= (MAP2K7 or MAP2K4) and not (DUSP1)
DUSP1 *= (MSK_f or MAPK14)
MAPK14 *= (MAP2K3 or MAP2K4) and not (DUSP1)
MAP2K3 *= (MAP3K5 or MAP3K7)
MAP3K5 *= not (AKT_f)
AKT_f *= (mTORC2_c or PDPK1)
TAB_f *= not (MAPK14)
MSK_f *= (ERK_f or MAPK14)
RTPK_f *= (MMP_f or RTPK_g) and not (MEK_f or MAPK14)
RTPK_g *= (FOXO_f)
MEK_f *= (RAF_f or MAP3K8) and not (ERK_f)
SHC1 *= (RTPK_f) and not (PTEN)
PTEN *= (PTEN_g) and not (GSK3_f)
GRB2 *= (SHC1)
SOS1 *= (GRB2) and not (ERK_f)
ERK_f *= (MEK_f) and not (DUSP6)
KRAS *= (PTPN11 or SOS1)
PTPN11 *= (GAB_f)
RAF_f *= (KRAS) and not (RHEB or ERK_f or AKT_f)
RHEB *= not (TSC_f)
MAP3K8 *= (IKBKB)
DUSP6 *= (ERK_f or mTORC1_c)
mTORC1_c *= (RHEB or RSK_f) and not (AKT1S1)
GAB_f *= (GRB2) and not (ERK_f)
RSK_f *= (ERK_f or PDPK1)
PDPK1 *= (PIK3CA) and not (PTEN)
PIK3CA *= (KRAS or GAB_f or IRS1)
IRS1 *= not (IKBKB or S6K_f or ERK_f)
S6K_f *= (PDPK1 or mTORC1_c)
IKBKB *= (MAP3K7) and not (TP53)
PTEN_g *= (EGR1)
GSK3_f *= not (ERK_f or AKT_f or MAPK14 or DVL_f or LRP_f or S6K_f or RSK_f)
TSC_f *= (GSK3_f) and not (IKBKB or ERK_f or AKT_f or RSK_f)
TP53 *= (MAPK14) and not (MDM2)
AKT1S1 *= not (AKT_f)
MDM2 *= (MDM2_g or AKT_f) and not (S6K_f)
MDM2_g *= (TP53 or NFKB_f)
BAX *= (TP53)
CYCS *= (BAX) and not (BCL2)
BCL2 *= not (BAD)
BAD *= not (AKT_f or RSK_f)
NLK *= (MAP3K7)
CASP9 *= (CYCS)
CASP8 *= not (CFLAR)
CFLAR *= (AKT_f) and not (ITCH)
ITCH *= (JNK_f)
FZD_f *= not (SFRP1)
SFRP1 *= (SFRP1_g)
LRP_f *= (FZD_f or JNK_f or ERK_f or MAPK14) and not (DKK_f)
DKK_f *= (DKK_g)
AXIN1 *= not (LRP_f)
CK1_f *= not (LRP_f)
BTRC *= (GSK3_f or CK1_f or AXIN1)
CTNNB1 *= (CHUK) and not (BTRC)
CHUK *= (AKT_f)
TCF7_f *= (CTNNB1) and not (NLK)
EGR1 *= not (TCF7_f)
DKK_g *= (TCF7_f) and not (MYC)
SFRP1_g *= not (MYC)
NFKB_f *= (IKBKB or CHUK or MSK_f)
LEF *= (CTNNB1)
MMP_f *= (LEF)
Initialize Boolean Model from .bnet file#
See more about the BoolNet format: CoLoMoTo formats and PyBoolNet Docs.
Creating a BooleanModel, that can be initialized:
file(required): The path to the.bnetfile.model_name: The name of the model.mutation_type: There are 3 options:topology,mixed,balanced. The default is balanced.attractor_tool: There are 2 options:mpbnorpyboolnetThe default ismpbn.attractor_type:There are 2 options:stable_statesortrapspaces. The default isstable_states.
equations_bnet = 'network.bnet'
boolean_model_bnet = BooleanModel(file=equations_bnet, model_name='test2',
mutation_type='balanced', attractor_tool='mpbn', attractor_type='trapspaces')
2024-12-05 23:41:39,714 - INFO - Boolean Model from .bnet file is created.
Run Train and Predict with executor#
Required Parameters:
BooleanModel,ModelOutputs,PerturbationsGenetic Algorithm Arguments (
ga_args)Evolution Arguments (
ev_args)Observed Synergy Scores (
observed_synergy_scores)
Optional Parameters:
Training Data
Initialize Train and Predict#
To run training and prediction, have to set up train_params and predict_params, which define how the executor handles model training and simulations. Below, you can see how to initialize these parameters.
1. train_params#
This dictionary holds the configuration and input data necessary for training Boolean Models.
Below is a detailed overview of its parameters:
boolean_model: The initialBooleanModelinstance that serves as the starting point for training.model_outputs: An instance ofModelOutputs.training_data(optional): An instance ofTrainingData.ga_args: A dictionary of arguments and configurations for the genetic algorithm.ev_args: A dictionary of arguments for the evolutionary strategy settings. It runs multiple times the genetic algorithm.save_best_models(optional): Whether to save the models with the best fitness score to disk. By defult,False.save_path(optional): The path where the best models will be saved ifsave_best_modelsisTrue. (e.g.'./models').
Arguments for the Genetic Algorithm (ga_args)#
The Evolution pipeline uses the PyGAD Genetic Algoritm . For more information about the PyGAD.GA initialization click here .
ga_args = {
'num_generations': 20,
'num_parents_mating': 3,
'mutation_num_genes': 8,
'fitness_batch_size': 20, # should be the same number as the num_generations
'crossover_type': 'single_point',
'mutation_type': 'random',
'keep_elitism': 6,
# 'stop_criteria': 'reach_95'
}
Arguments for the Evolution (ev_args)#
num_best_solutions: Number of the best solutions per Evolution run.num_of_runs: Number of running the Evolutionnum_of_cores: Maximum number of cores for calculationsnum_of_init_mutation: Number of mutated genes in the initial population.
ev_args = {
'num_best_solutions': 3,
'num_of_runs': 50,
'num_of_cores': 4,
'num_of_init_mutation': 20
}
Init train_params#
train_params = {
'boolean_model': boolean_model_bnet,
'model_outputs': model_outputs,
'training_data': training_data,
'ga_args': ga_args,
'ev_args': ev_args,
'save_best_models': False,
# 'save_path': './models'
}
2. predict_params#
This dictionary holds the necessary configurations for the predict function, which simulates perturbed models and predicts drug synergies.
Below is a detailed overview of its parameters:
perturbations: An instance ofPerturbation.model_outputs: An instance ofModelOutputs.observed_synergy_scores: A list of observed synergy scores used to plot and evaluate the predictive accuracy of the models.synergy_method: The method used to calculate synergy scores. Options:'bliss'(Bliss Independence) or'hsa'(Highest Single Agent). By default,'bliss'run_parallel(optional): Whether to run it paralelly or serially, By default,True.plot_roc_pr_curves(optional): Whether to plot the ROC and PR Curves when the predction finishes. By default,True.save_predictions(optional): Whether to save the predictions to disk after the simulations. By default,False.save_path(optional): Specifies the path to save the predictions ifsave_predictionsisTrue(e.g.'./predictions').For running only predict on previously trained models from disk:
model_directory: Path to a directory containing pre-trained models to be loaded ifbest_boolean_modelsis not provided.(e.g.,'./models/models_2024_11_06_1130').attractor_tool: Specifies the tool used for attractor computation if the models are loaded frommodel_directory(e.g.'mpbn').attractor_type: Specifies the tool used for attractor computation if the models are loaded frommodel_directory(e.g.'stable_states').cores: Number of CPU cores to use for parallel processing.(e.g.4for using four CPU cores).
For running only predict on previously trained models during runtime:
best_boolean_models: retults of train (e.g. models = train(boolean_model=boolean_model_bnet, model_outputs=model_outputs, training_data=training_data, ga_args=ga_args, ev_args=ev_args)cores: Number of CPU cores to use for parallel processing.(e.g.4for using four CPU cores).
Observed Synergy Scores#
List of observed synergy scores corresponding to the perturbations applied, used to evaluate model predictions.
Initialization: From a list containing observed synergy scores.
observed_synergy_scores = ["PI-PD", "PI-5Z", "PD-AK", "AK-5Z"]
Init predict_param#
predict_params = {
'perturbations': perturbations,
'model_outputs': model_outputs,
'observed_synergy_scores': observed_synergy_scores,
'synergy_method': 'bliss',
'plot_roc_pr_curves': True,
'save_predictions': False,
# 'cores':3,
# 'save_path': './predictions',
# 'model_directory': './models/example_models',
# 'attractor_tool': 'mpbn',
# 'attractor_type': 'stable_states'
}
Run Train and Predict#
from pydruglogics.execution.Executor import execute, train, predict
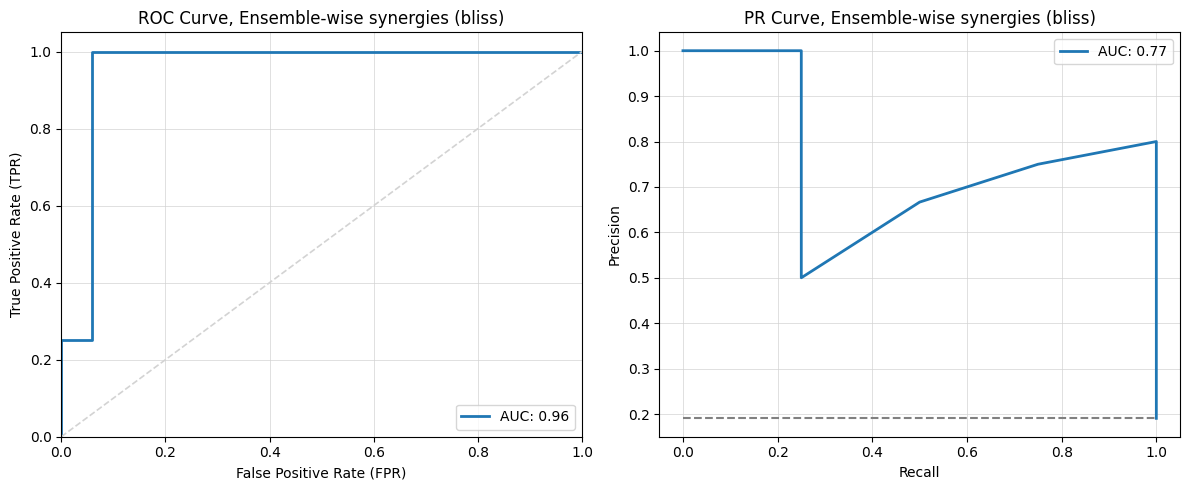
execute(train_params=train_params, predict_params=predict_params)
2024-12-05 23:41:39,787 - INFO - Train started...
2024-12-05 23:41:48,221 - INFO - Training finished.
2024-12-05 23:41:48,222 - INFO - Train completed in 8.44 seconds.
2024-12-05 23:41:48,223 - INFO - Predict started...
2024-12-05 23:42:00,944 - INFO -
Synergy scores (bliss):
2024-12-05 23:42:00,945 - INFO - PI-PD: -0.08418595679012342
2024-12-05 23:42:00,946 - INFO - PI-CT: 0.002967592592592605
2024-12-05 23:42:00,947 - INFO - PI-BI: 0.02335030864197518
2024-12-05 23:42:00,947 - INFO - PI-PK: 0.020891203703703565
2024-12-05 23:42:00,947 - INFO - PI-AK: 0.06716280864197521
2024-12-05 23:42:00,948 - INFO - PI-5Z: -0.036652777777777645
2024-12-05 23:42:00,948 - INFO - PD-CT: 0.0032314814814814463
2024-12-05 23:42:00,949 - INFO - PD-BI: 0.0024660493827158847
2024-12-05 23:42:00,949 - INFO - PD-PK: 0.1255787037037036
2024-12-05 23:42:00,949 - INFO - PD-AK: -0.043075617283950596
2024-12-05 23:42:00,950 - INFO - PD-5Z: 0.00913888888888903
2024-12-05 23:42:00,950 - INFO - CT-BI: -5.925925925931441e-05
2024-12-05 23:42:00,950 - INFO - CT-PK: 0.0023055555555555607
2024-12-05 23:42:00,951 - INFO - CT-AK: 0.0025129629629628836
2024-12-05 23:42:00,951 - INFO - CT-5Z: 0.0032999999999999696
2024-12-05 23:42:00,951 - INFO - BI-PK: -0.07506481481481486
2024-12-05 23:42:00,952 - INFO - BI-AK: 0.05955987654320993
2024-12-05 23:42:00,952 - INFO - BI-5Z: 0.083988888888889
2024-12-05 23:42:00,952 - INFO - PK-AK: 0.05078240740740736
2024-12-05 23:42:00,953 - INFO - PK-5Z: 0.18247222222222226
2024-12-05 23:42:00,953 - INFO - AK-5Z: -0.06634999999999991
2024-12-05 23:42:00,973 - INFO - Predicted Data with Observed Synergies for Model 1:
2024-12-05 23:42:00,973 - INFO - perturbation synergy_score observed
0 PI-PD 0.084186 1
1 BI-PK 0.075065 0
2 AK-5Z 0.066350 1
3 PD-AK 0.043076 1
4 PI-5Z 0.036653 1
5 CT-BI 0.000059 0
6 CT-PK -0.002306 0
7 PD-BI -0.002466 0
8 CT-AK -0.002513 0
9 PI-CT -0.002968 0
10 PD-CT -0.003231 0
11 CT-5Z -0.003300 0
12 PD-5Z -0.009139 0
13 PI-PK -0.020891 0
14 PI-BI -0.023350 0
15 PK-AK -0.050782 0
16 BI-AK -0.059560 0
17 PI-AK -0.067163 0
18 BI-5Z -0.083989 0
19 PD-PK -0.125579 0
20 PK-5Z -0.182472 0
2024-12-05 23:42:01,167 - INFO - Predict completed in 12.94 seconds.
2024-12-05 23:42:01,168 - INFO - Total runtime for training and prediction: 21.38 seconds
Alternatively, train and predict can be run separately:
# execute(train_params=train_params)
Note: If you would like to run the predict process but the train process has not been previously executed, it is necessary to load the Boolean Models from a .bnet file and add the path to the model_directory. In this case attractor_tool and attractor_type must be provided.
Run Statistics#
Train Boolean Models#
Boolean Models calibrated to steady state response.
best_boolean_models_calibrated = train(boolean_model=boolean_model_bnet, model_outputs=model_outputs, training_data=training_data,
ga_args=ga_args, ev_args=ev_args)
2024-12-05 23:42:01,187 - INFO - Train started...
2024-12-05 23:42:09,851 - INFO - Training finished.
2024-12-05 23:42:09,852 - INFO - Train completed in 8.66 seconds.
Boolean Models match to proliferation profile.
best_boolean_models_random = train(boolean_model=boolean_model_bnet, model_outputs=model_outputs, ga_args=ga_args, ev_args=ev_args)
2024-12-05 23:42:09,860 - INFO - Train started...
2024-12-05 23:42:09,861 - INFO - Training data initialized from list.
2024-12-05 23:42:12,241 - INFO - Training finished.
2024-12-05 23:42:12,242 - INFO - Train completed in 2.38 seconds.
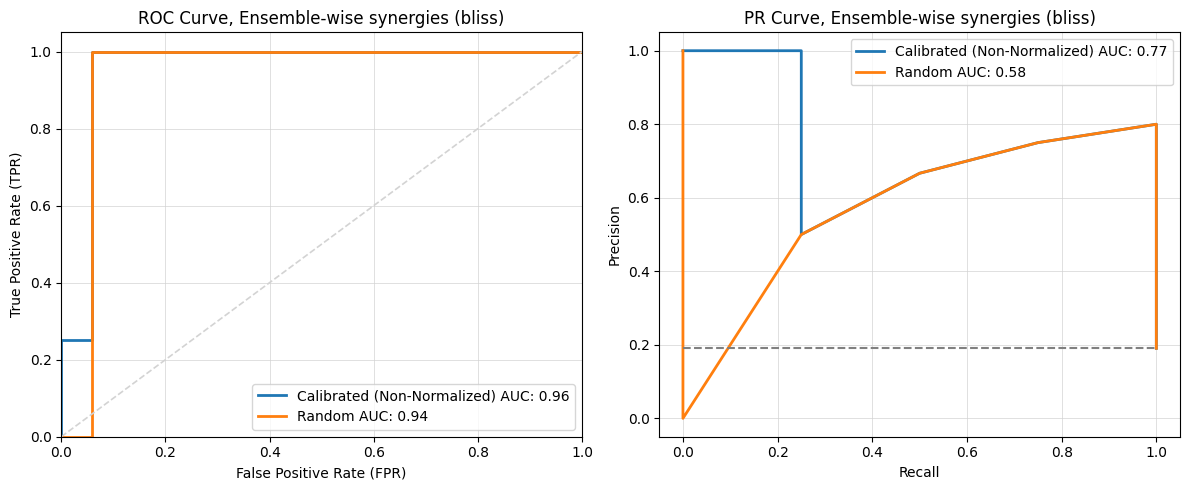
Compare Boolean Models trained by calibrated and random data#
The compare_two_simulations function compares the predictive performance of two sets of evolved Boolean models by plotting ROC and PR curves.
Parameters:#
boolean_models1: A list of the best Boolean models from the first run.boolean_models2: A list of the best Boolean models from the second run.observed_synergy_scores: A reference list of observed synergy scores used to evaluate the predictive performance of the models.model_outputs: An instance ofModelOutputs.perturbations: An instance ofPerturbation.synergy_method(optional): The method for assessing synergy. Options:'hsa'(Highest Single Agent) or'bliss'(Bliss Independence). By default,'bliss'.label1(optional): A custom label for the first set of evolution results shown in the plots. By default,'Models 1'.label2(optional): A custom label for the second set of evolution results shown in the plots. By default,'Models 2'.normalized(optional): Whether or not to normalize the synergy scores of the first set. By default,True.plot_pr_roc_curves(optional): Whether or not to plot ROC and PR Curves. By default,True.save_result(optional): Whether or not to save the results to file. By default,True.
from pydruglogics.statistics.Statistics import compare_two_simulations
compare_two_simulations(boolean_models1=best_boolean_models_calibrated, boolean_models2=best_boolean_models_random, observed_synergy_scores=observed_synergy_scores,
model_outputs=model_outputs,perturbations=perturbations, synergy_method='bliss',label1='Calibrated (Non-Normalized)',
label2='Random', normalized=False, plot=True, save_result=False)
2024-12-05 23:42:25,285 - INFO -
Synergy scores (bliss):
2024-12-05 23:42:25,286 - INFO - PI-PD: -0.08418595679012342
2024-12-05 23:42:25,287 - INFO - PI-CT: 0.002967592592592605
2024-12-05 23:42:25,288 - INFO - PI-BI: 0.02335030864197518
2024-12-05 23:42:25,288 - INFO - PI-PK: 0.020891203703703565
2024-12-05 23:42:25,289 - INFO - PI-AK: 0.06716280864197521
2024-12-05 23:42:25,289 - INFO - PI-5Z: -0.036652777777777645
2024-12-05 23:42:25,290 - INFO - PD-CT: 0.0032314814814814463
2024-12-05 23:42:25,290 - INFO - PD-BI: 0.0024660493827158847
2024-12-05 23:42:25,290 - INFO - PD-PK: 0.1255787037037036
2024-12-05 23:42:25,291 - INFO - PD-AK: -0.043075617283950596
2024-12-05 23:42:25,291 - INFO - PD-5Z: 0.00913888888888903
2024-12-05 23:42:25,291 - INFO - CT-BI: -5.925925925931441e-05
2024-12-05 23:42:25,292 - INFO - CT-PK: 0.0023055555555555607
2024-12-05 23:42:25,292 - INFO - CT-AK: 0.0025129629629628836
2024-12-05 23:42:25,292 - INFO - CT-5Z: 0.0032999999999999696
2024-12-05 23:42:25,292 - INFO - BI-PK: -0.07506481481481486
2024-12-05 23:42:25,292 - INFO - BI-AK: 0.05955987654320993
2024-12-05 23:42:25,293 - INFO - BI-5Z: 0.083988888888889
2024-12-05 23:42:25,293 - INFO - PK-AK: 0.05078240740740736
2024-12-05 23:42:25,293 - INFO - PK-5Z: 0.18247222222222226
2024-12-05 23:42:25,293 - INFO - AK-5Z: -0.06634999999999991
2024-12-05 23:42:36,667 - INFO -
Synergy scores (bliss):
2024-12-05 23:42:36,668 - INFO - PI-PD: -0.10488888888888914
2024-12-05 23:42:36,669 - INFO - PI-CT: 0.0
2024-12-05 23:42:36,669 - INFO - PI-BI: 0.04377777777777758
2024-12-05 23:42:36,669 - INFO - PI-PK: 0.08273333333333321
2024-12-05 23:42:36,670 - INFO - PI-AK: 0.1151777777777776
2024-12-05 23:42:36,670 - INFO - PI-5Z: -0.021911111111111325
2024-12-05 23:42:36,670 - INFO - PD-CT: 0.0
2024-12-05 23:42:36,671 - INFO - PD-BI: 0.023796296296296204
2024-12-05 23:42:36,671 - INFO - PD-PK: 0.18807407407407395
2024-12-05 23:42:36,671 - INFO - PD-AK: -0.023129629629629722
2024-12-05 23:42:36,671 - INFO - PD-5Z: 0.0032962962962962417
2024-12-05 23:42:36,671 - INFO - CT-BI: 0.0
2024-12-05 23:42:36,672 - INFO - CT-PK: 0.0
2024-12-05 23:42:36,672 - INFO - CT-AK: 0.0
2024-12-05 23:42:36,672 - INFO - CT-5Z: 0.0
2024-12-05 23:42:36,672 - INFO - BI-PK: -0.13427777777777794
2024-12-05 23:42:36,673 - INFO - BI-AK: 0.08134722222222213
2024-12-05 23:42:36,673 - INFO - BI-5Z: 0.05813888888888874
2024-12-05 23:42:36,673 - INFO - PK-AK: 0.10301111111111105
2024-12-05 23:42:36,673 - INFO - PK-5Z: 0.08075555555555558
2024-12-05 23:42:36,674 - INFO - AK-5Z: -0.03427222222222237
2024-12-05 23:42:36,689 - INFO - Predicted Data with Observed Synergies for Calibrated (Non-Normalized):
2024-12-05 23:42:36,690 - INFO - perturbation synergy_score observed
0 PI-PD 0.084186 1
1 BI-PK 0.075065 0
2 AK-5Z 0.066350 1
3 PD-AK 0.043076 1
4 PI-5Z 0.036653 1
5 CT-BI 0.000059 0
6 CT-PK -0.002306 0
7 PD-BI -0.002466 0
8 CT-AK -0.002513 0
9 PI-CT -0.002968 0
10 PD-CT -0.003231 0
11 CT-5Z -0.003300 0
12 PD-5Z -0.009139 0
13 PI-PK -0.020891 0
14 PI-BI -0.023350 0
15 PK-AK -0.050782 0
16 BI-AK -0.059560 0
17 PI-AK -0.067163 0
18 BI-5Z -0.083989 0
19 PD-PK -0.125579 0
20 PK-5Z -0.182472 0
2024-12-05 23:42:36,693 - INFO - Predicted Data with Observed Synergies for Random:
2024-12-05 23:42:36,693 - INFO - perturbation synergy_score observed
0 BI-PK 0.134278 0
1 PI-PD 0.104889 1
2 AK-5Z 0.034272 1
3 PD-AK 0.023130 1
4 PI-5Z 0.021911 1
5 CT-AK -0.000000 0
6 CT-5Z -0.000000 0
7 CT-BI -0.000000 0
8 PI-CT -0.000000 0
9 CT-PK -0.000000 0
10 PD-CT -0.000000 0
11 PD-5Z -0.003296 0
12 PD-BI -0.023796 0
13 PI-BI -0.043778 0
14 BI-5Z -0.058139 0
15 PK-5Z -0.080756 0
16 BI-AK -0.081347 0
17 PI-PK -0.082733 0
18 PK-AK -0.103011 0
19 PI-AK -0.115178 0
20 PD-PK -0.188074 0
Sampling#
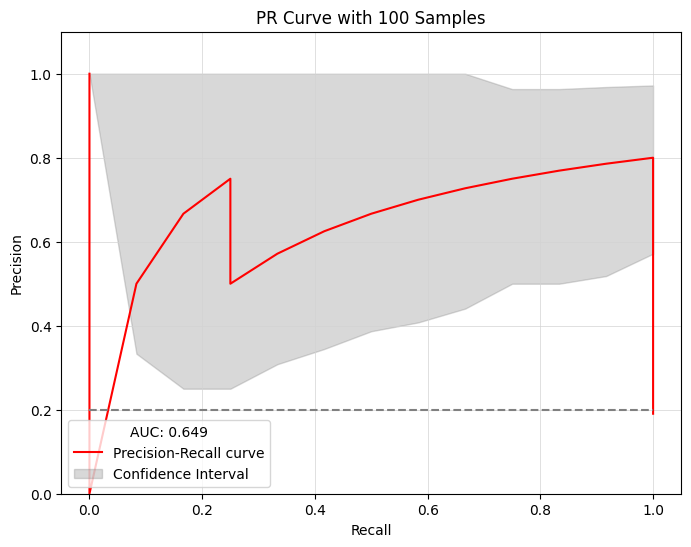
The sampling_with_ci function performs sampling on a list of Boolean models, calculates confidence intervals, and plots a Precision-Recall (PR) curve to assess the predictive performance in identifying synergistic drug combinations.
Parameters:#
boolean_models: A list ofBooleanModelinstances used for the sampling.observed_synergy_scores: A list of observed synergy scores.model_outputs: An instance ofModelOutputs.perturbations: An instance ofPerturbation.synergy_method(optional): The method for assessing synergy. Options:'hsa'(Highest Single Agent) or'bliss'(Bliss Independence). By default,'bliss'.repeat_time(optional): The number of times to repeat the sampling process. By defualt,10.sub_ratio(optional): The proportion of models to sample in each iteration. By defualt,0.8.boot_n(optional): The number of bootstrap iterations for calculating confidence intervals. By defualt,1000.confidence_level(optional): The confidence level used for the interval calculations. By defualt,0.9.plot(optional): Whether to plot the PR curve. By defualt,True.plot_discrete(optional): Whether to plot discrete points instead of continuous line on the PR curve. By defualt,False.save_result(optional): Whether to save the results. By defualt,True.with_seeds(optional): Whether to use a fixed seed for reproducibility. By defualt,True.seeds(optional): The seed value to ensure reproducibility. By defualt,42.
from pydruglogics.statistics.Statistics import sampling_with_ci
sampling_with_ci(boolean_models=best_boolean_models_calibrated, observed_synergy_scores=observed_synergy_scores, model_outputs=model_outputs,
perturbations=perturbations,synergy_method='bliss', repeat_time=3, sub_ratio=0.8, boot_n=100, confidence_level= 0.95,
plot=True, plot_discrete=False, save_result=False, with_seeds=True, seeds=42)
2024-12-05 23:42:46,170 - INFO -
Synergy scores (bliss):
2024-12-05 23:42:46,170 - INFO - PI-PD: -0.08004557291666681
2024-12-05 23:42:46,171 - INFO - PI-CT: 0.0037254050925926707
2024-12-05 23:42:46,172 - INFO - PI-BI: 0.02237533757716037
2024-12-05 23:42:46,173 - INFO - PI-PK: 0.016493055555555358
2024-12-05 23:42:46,173 - INFO - PI-AK: 0.06550564236111112
2024-12-05 23:42:46,174 - INFO - PI-5Z: -0.0360460069444446
2024-12-05 23:42:46,174 - INFO - PD-CT: 0.004036458333333437
2024-12-05 23:42:46,175 - INFO - PD-BI: 0.0037109375000000666
2024-12-05 23:42:46,176 - INFO - PD-PK: 0.11840277777777775
2024-12-05 23:42:46,176 - INFO - PD-AK: -0.041644965277777724
2024-12-05 23:42:46,177 - INFO - PD-5Z: 0.011414930555555647
2024-12-05 23:42:46,177 - INFO - CT-BI: -8.391203703683736e-05
2024-12-05 23:42:46,178 - INFO - CT-PK: 0.0029166666666667895
2024-12-05 23:42:46,178 - INFO - CT-AK: 0.0031510416666666874
2024-12-05 23:42:46,178 - INFO - CT-5Z: 0.004114583333333477
2024-12-05 23:42:46,179 - INFO - BI-PK: -0.083125
2024-12-05 23:42:46,179 - INFO - BI-AK: 0.059500868055555545
2024-12-05 23:42:46,182 - INFO - BI-5Z: 0.08759548611111134
2024-12-05 23:42:46,182 - INFO - PK-AK: 0.044930555555555474
2024-12-05 23:42:46,183 - INFO - PK-5Z: 0.18166666666666664
2024-12-05 23:42:46,183 - INFO - AK-5Z: -0.06763020833333322
2024-12-05 23:42:55,315 - INFO -
Synergy scores (bliss):
2024-12-05 23:42:55,316 - INFO - PI-PD: -0.08228081597222237
2024-12-05 23:42:55,316 - INFO - PI-CT: 0.003705150462963047
2024-12-05 23:42:55,316 - INFO - PI-BI: 0.02233603395061723
2024-12-05 23:42:55,317 - INFO - PI-PK: 0.017199315200617238
2024-12-05 23:42:55,317 - INFO - PI-AK: 0.06817973572530855
2024-12-05 23:42:55,318 - INFO - PI-5Z: -0.03857687114197528
2024-12-05 23:42:55,318 - INFO - PD-CT: 0.004036458333333437
2024-12-05 23:42:55,318 - INFO - PD-BI: -0.0005208333333334147
2024-12-05 23:42:55,319 - INFO - PD-PK: 0.11942274305555556
2024-12-05 23:42:55,319 - INFO - PD-AK: -0.04242621527777779
2024-12-05 23:42:55,319 - INFO - PD-5Z: 0.009418402777777835
2024-12-05 23:42:55,320 - INFO - CT-BI: -9.259259259253305e-05
2024-12-05 23:42:55,320 - INFO - CT-PK: 0.0028674768518519578
2024-12-05 23:42:55,320 - INFO - CT-AK: 0.0031394675925926485
2024-12-05 23:42:55,321 - INFO - CT-5Z: 0.004126157407407516
2024-12-05 23:42:55,321 - INFO - BI-PK: -0.07249614197530874
2024-12-05 23:42:55,322 - INFO - BI-AK: 0.05869984567901232
2024-12-05 23:42:55,322 - INFO - BI-5Z: 0.08786265432098761
2024-12-05 23:42:55,322 - INFO - PK-AK: 0.04952015817901234
2024-12-05 23:42:55,323 - INFO - PK-5Z: 0.18655189043209897
2024-12-05 23:42:55,323 - INFO - AK-5Z: -0.06559124228395063
2024-12-05 23:43:04,572 - INFO -
Synergy scores (bliss):
2024-12-05 23:43:04,573 - INFO - PI-PD: -0.07469666280864196
2024-12-05 23:43:04,574 - INFO - PI-CT: 0.0037080439814816124
2024-12-05 23:43:04,574 - INFO - PI-BI: 0.025728684413580227
2024-12-05 23:43:04,574 - INFO - PI-PK: 0.022124083719135768
2024-12-05 23:43:04,575 - INFO - PI-AK: 0.06542679398148155
2024-12-05 23:43:04,575 - INFO - PI-5Z: -0.03262586805555545
2024-12-05 23:43:04,575 - INFO - PD-CT: 0.004033564814814872
2024-12-05 23:43:04,575 - INFO - PD-BI: -0.0020273919753087677
2024-12-05 23:43:04,576 - INFO - PD-PK: 0.12402295524691354
2024-12-05 23:43:04,576 - INFO - PD-AK: -0.035896990740740486
2024-12-05 23:43:04,576 - INFO - PD-5Z: 0.011406250000000173
2024-12-05 23:43:04,576 - INFO - CT-BI: -9.837962962944147e-05
2024-12-05 23:43:04,576 - INFO - CT-PK: 0.0028616898148148273
2024-12-05 23:43:04,576 - INFO - CT-AK: 0.003107638888888986
2024-12-05 23:43:04,576 - INFO - CT-5Z: 0.004114583333333477
2024-12-05 23:43:04,577 - INFO - BI-PK: -0.0682166280864197
2024-12-05 23:43:04,577 - INFO - BI-AK: 0.06309606481481478
2024-12-05 23:43:04,577 - INFO - BI-5Z: 0.08963541666666663
2024-12-05 23:43:04,577 - INFO - PK-AK: 0.05581307870370378
2024-12-05 23:43:04,577 - INFO - PK-5Z: 0.1815017361111111
2024-12-05 23:43:04,577 - INFO - AK-5Z: -0.0587326388888888