Predict mutations with Pint, refine with MaBoSS#
This notebook shows a possible way to combine formal prediction of mutation performed by Pint with the quantiative evaluation of attractors reachability with MaBoSS.
A detailed use case of such an analysis can be found in the notebook “Usecase - Mutations enabling tumour invasion”.
Model#
We load a model of cell fate decision model from CellCollective using bioLQM:
import biolqm
lqm = biolqm.load("http://ginsim.org/sites/default/files/SuppMat_Model_Master_Model.zginml")
Downloading ‘http://ginsim.org/sites/default/files/SuppMat_Model_Master_Model.zginml’
Wild-type simulation with MaBoSS#
We convert the model to MaBoSS, and configure the simulation
import maboss
wt_sim = biolqm.to_maboss(lqm)
wt_sim.network.set_output(('Metastasis', 'Migration', 'Invasion', 'Apoptosis', 'CellCycleArrest'))
wt_sim.network.set_istate("ECMicroenv", [0, 1]) # ECM is active
wt_sim.network.set_istate("DNAdamage", [0.5, 0.5]) # DNAdamage can start either active or inactive
wt_sim.update_parameters(max_time=50)
We perform the simulation with MaBoSS, this can take several seconds.
wt_res = wt_sim.run()
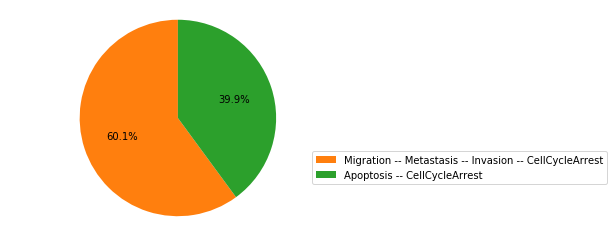
We plot the distribution of attractors at the end of the simulations:
wt_res.plot_piechart()
Mutation prediction with Pint#
Now, we use Pint to predict mutations which remove any possible activation of apoptosis.
import pypint
m = biolqm.to_pint(lqm)
m.initial_state["ECMicroenv"] = 1
m.initial_state["DNAdamage"] = {0,1}
mutants = m.oneshot_mutations_for_cut("Apoptosis=1", exclude={"ECMicroenv", "DNAdamage"})
mutants
This computation is an under-approximation: returned mutations are all valid, but they may be non-minimal, and some solutions may be missed.
Limiting solutions to mutations of at most 5 automata. Use maxsize argument to change.
[{'ZEB2': 1},
{'AKT1': 1},
{'AKT2': 1},
{'ERK': 1},
{'SNAI2': 1, 'ZEB1': 1, 'NICD': 1},
{'SNAI2': 1, 'ZEB1': 1, 'p63': 0},
{'SNAI2': 1, 'ZEB1': 1, 'miR203': 1},
{'SNAI2': 1, 'NICD': 1, 'p73': 0},
{'SNAI2': 1, 'p63': 0, 'p73': 0},
{'SNAI2': 1, 'p73': 0, 'miR203': 1},
{'ZEB1': 1, 'NICD': 1, 'p53': 0},
{'ZEB1': 1, 'p63': 0, 'p53': 0},
{'ZEB1': 1, 'p53': 0, 'miR203': 1},
{'NICD': 1, 'p53': 0, 'p73': 0},
{'p63': 0, 'p53': 0, 'p73': 0},
{'p53': 0, 'p73': 0, 'miR203': 1}]
Each returned solution gives a combination of mutations which are guaranteed to remove the capability to activate apoptosis, even transiently.
Collecting experiments#
Among the results returned by Pint, we want to try any potential double-mutant that may be sufficient to remove the reachability of a stable apoptosis.
We use Python standard library functions to compute all the couple of mutations in each predicted mutations, and then merge these couples in a single set of candidate double-mutants.
from itertools import combinations
from functools import reduce
mutant_combinations = [combinations(m.items(), 2) for m in mutants if len(m) >= 2]
candidates = reduce(set.union, mutant_combinations, set())
candidates
{(('NICD', 1), ('p53', 0)),
(('NICD', 1), ('p73', 0)),
(('SNAI2', 1), ('NICD', 1)),
(('SNAI2', 1), ('ZEB1', 1)),
(('SNAI2', 1), ('miR203', 1)),
(('SNAI2', 1), ('p63', 0)),
(('SNAI2', 1), ('p73', 0)),
(('ZEB1', 1), ('NICD', 1)),
(('ZEB1', 1), ('miR203', 1)),
(('ZEB1', 1), ('p53', 0)),
(('ZEB1', 1), ('p63', 0)),
(('p53', 0), ('miR203', 1)),
(('p53', 0), ('p73', 0)),
(('p63', 0), ('p53', 0)),
(('p63', 0), ('p73', 0)),
(('p73', 0), ('miR203', 1))}
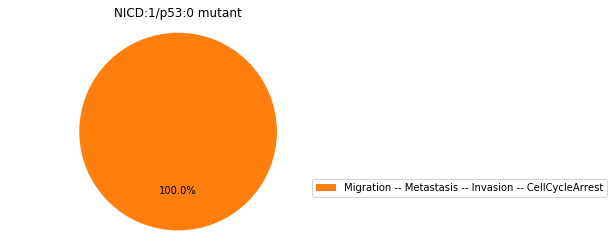
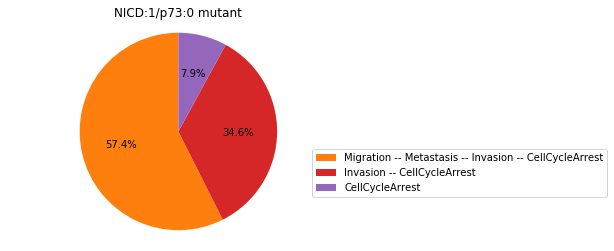
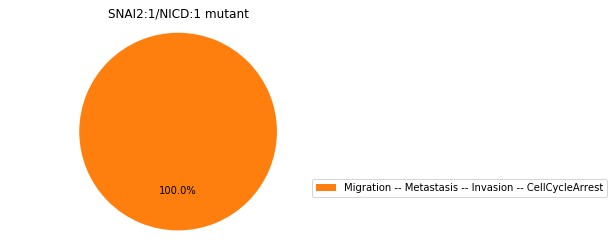
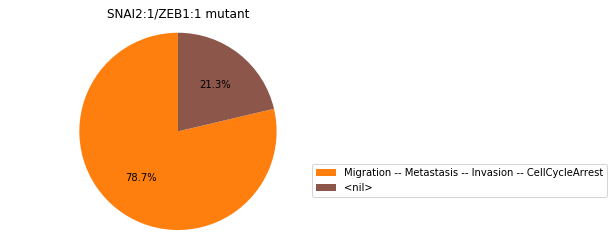
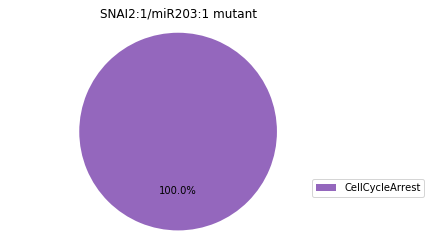
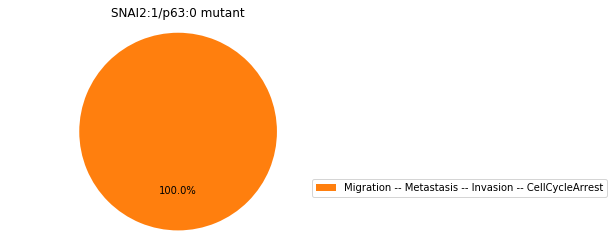
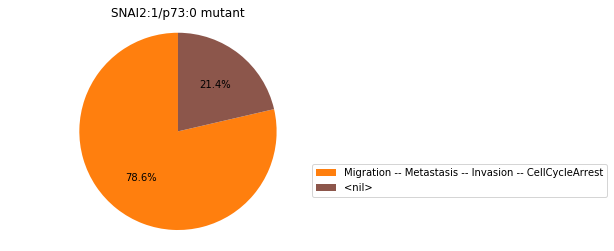
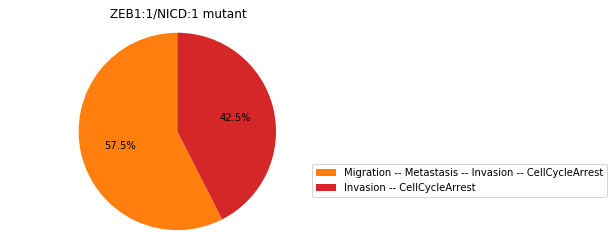
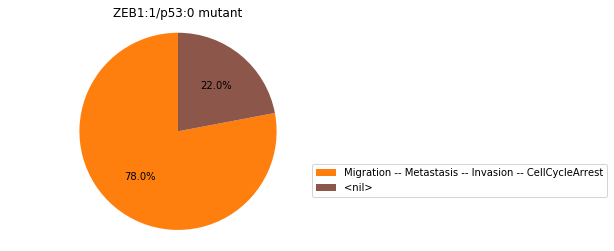
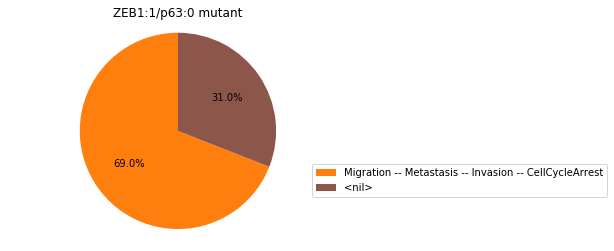
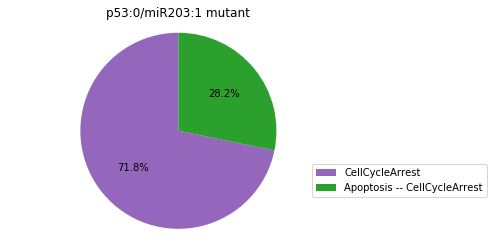
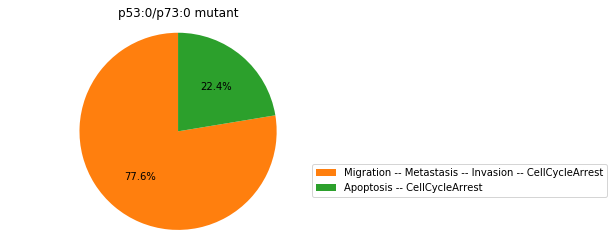
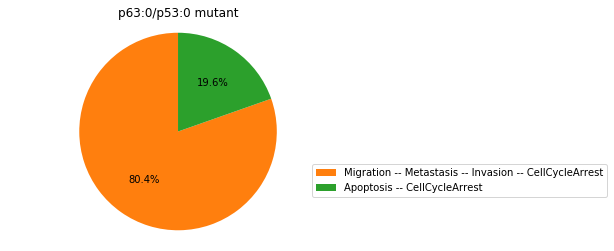
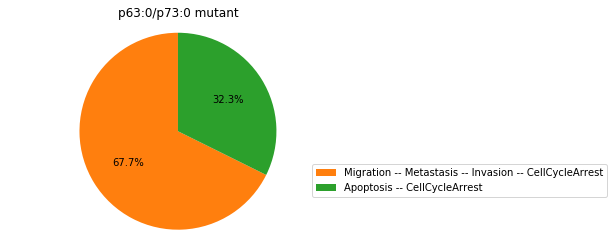
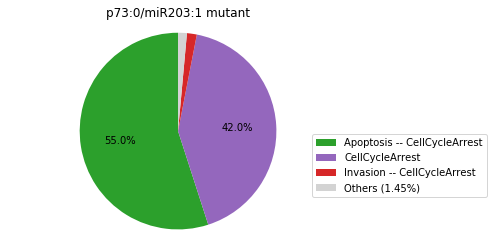
Double-mutant experiments with MaBoSS#
For each candidate double-mutant, we copy the MaBoSS wild-type model, apply the corresponding mutation and run the simulations. As there are 16 candidates to evaluate, the overall execution can take several minutes.
import matplotlib.pyplot as plt # for customizing the plots
for mutant in sorted(candidates):
mut_sim = wt_sim.copy()
for (node, value) in mutant:
mut_sim.mutate(node, "ON" if value else "OFF")
mut_res = mut_sim.run()
mut_res.plot_piechart(embed_labels=False, autopct=4)
mutant_name = "/".join(["%s:%s"%m for m in mutant])
plt.title("%s mutant" % mutant_name)