BooN basic features#
See https://franck-delaplace.github.io/BooN/ for documentation.
import matplotlib.pyplot as plt
import networkx as nx
from tabulate import tabulate
from boon import *
from boon.logic import *
Boolean network definition#
from sympy.abc import w, x, y, z, v
boon = BooN({x: y, y: x & z, z: w | ~x & v | y, w: x & y | ~x & z & ~w & ~v, v: ~v & w})
Variables of BooN:
print(boon.variables)
{x, v, w, y, z}
Print styles#
Logical (default):
print(boon)
x = y
v = w ∧ ¬v
w = (x ∧ y) ∨ (z ∧ ¬v ∧ ¬w ∧ ¬x)
y = x ∧ z
z = w ∨ y ∨ (v ∧ ¬x)
Sympy:
boon.style = SYMPY
print(boon)
x = y
v = w & ~v
w = (x & y) | (z & ~v & ~w & ~x)
y = x & z
z = w | y | (v & ~x)
Mathematica:
boon.style = MATHEMATICA
print(boon)
x = y
v = w && !v
w = (x && y) || (z && !v && !w && !x)
y = x && z
z = w || y || (v && !x)
# Default style is LOGICAL
boon.style = LOGICAL
Delete v#
boon.delete(v)
print(boon)
z = w ∨ y ∨ ¬x
w = (x ∧ y) ∨ (z ∧ ¬w ∧ ¬x)
y = x ∧ z
x = y
Renaming#
Rename w to v
boon.rename(w, v)
print(boon)
z = v ∨ y ∨ ¬x
v = (x ∧ y) ∨ (z ∧ ¬v ∧ ¬x)
y = x ∧ z
x = y
Stable states#
stable = boon.stable_states
tabulate(stable, headers='keys', tablefmt="html")
| z | v | y | x |
|---|---|---|---|
| True | True | True | True |
Stability constraints in logic:
prettyform(boon.stability_constraints())
'(x ⇔ y) ∧ (y ⇔ (x ∧ z)) ∧ (z ⇔ (v ∨ y ∨ ¬x)) ∧ (v ⇔ ((x ∧ y) ∨ (z ∧ ¬v ∧ ¬x)))'
Dynamics#
Asynchronous model#
# Define the datamodel of dynamics
M = boon.model()
# initialize figures
_, ax2 = plt.subplots()
ax2.axis('off')
boon.draw_model(M, pos=hypercube_layout(4), ax=ax2)
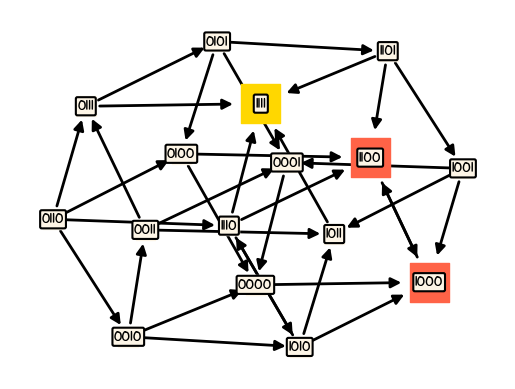
Synchronous model#
# Synchronous datamodel
MS = boon.model(mode=synchronous, self_loop=True)
_, ax3 = plt.subplots()
ax3.axis('off')
boon.draw_model(MS, pos=nx.shell_layout(MS), ax=ax3)
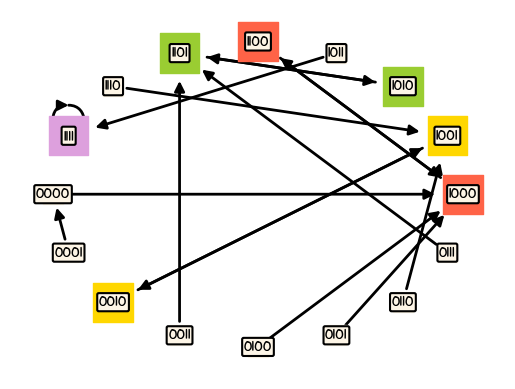
Equilibria#
eqs = boon.equilibria(model=MS)
for eq in eqs:
print(tabulate(eq, headers='keys'))
z v y x
---- ----- ----- -----
True False False False
True True False False
z v y x
----- ----- ----- -----
True False False True
False False True False
z v y x
---- ----- ----- -----
True False True False
True True False True
z v y x
---- ---- ---- ----
True True True True
Interaction graph#
IG = boon.interaction_graph
boon.pos = nx.circular_layout(IG)
_, ax1 = plt.subplots()
ax1.axis('off')
boon.draw_IG(IG, modular=True, ax=ax1)
<networkx.classes.digraph.DiGraph at 0x7f1903ebd8d0>
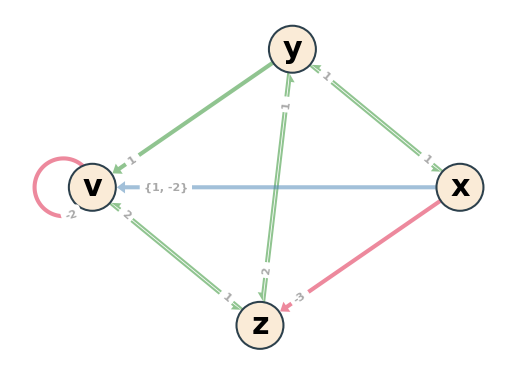
Retrieve BooN from the interaction graph#
boon.from_ig(IG)
print(boon)
z = v ∨ y ∨ ¬x
v = (x ∧ y) ∨ (z ∧ ¬v ∧ ¬x)
y = x ∧ z
x = y
Save and re-load#
boon.save("bn")
boon2 = BooN.load("bn")
print(boon2)
z = v ∨ y ∨ ¬x
v = (x ∧ y) ∨ (z ∧ ¬v ∧ ¬x)
y = x ∧ z
x = y
CNF conversion#
boon2.cnf()
print(boon2)
z = v ∨ y ∨ ¬x
v = (x ∨ z) ∧ (x ∨ ¬v) ∧ (y ∨ ¬x)
y = x ∧ z
x = y
DNF conversion#
boon2.dnf()
print(boon2)
z = v ∨ y ∨ ¬x
v = (x ∧ y) ∨ (z ∧ ¬v ∧ ¬x)
y = x ∧ z
x = y
Export and import from text file#
boon2.to_textfile('boolnet')
boon2 = BooN.from_textfile('boolnet.txt')
print(boon2)
z = v ∨ y ∨ ¬x
v = (x ∧ y) ∨ (z ∧ ¬v ∧ ¬x)
y = x ∧ z
x = y
Import from GINsim, and compute stable states#
import biolqm
This notebook has been executed using the docker image colomoto/colomoto-docker:2024-09-01
lqm = biolqm.load("http://ginsim.org/sites/default/files/Th_17.zginml")
Using local file Th_17.zginml
biolqm.save(lqm, "th17.bnet")
'th17.bnet'
b = BooN.from_textfile("th17.bnet")
print(b)
STAT6 = IL4R
Tbet_b2 = (STAT1_b1 ∧ STAT1_b2 ∧ Tbet_b1 ∧ ¬GATA3) ∨ (STAT1_b1 ∧ Tbet_b1 ∧ Tbet_b2 ∧ ¬STAT1_b2) ∨ (Tbet_b1 ∧ Tbet_b2 ∧ ¬GATA3 ∧ ¬STAT1_b1)
IL4 = GATA3 ∧ ¬STAT1_b1
IL12R = IL12 ∧ ¬STAT6
IFNbR = IFNb
IRAK = IL18R
SOCS1 = STAT1_b1 ∨ (Tbet_b1 ∧ ¬STAT1_b1)
IFNb = false
IFNg_b1 = (IFNg_b1 ∧ IFNg_b2) ∨ (Tbet_b1 ∧ ¬IFNg_b1) ∨ (IFNg_b1 ∧ Tbet_b1 ∧ ¬IFNg_b2) ∨ (STAT4 ∧ ¬IFNg_b1 ∧ ¬Tbet_b1) ∨ (IFNg_b1 ∧ STAT4 ∧ ¬IFNg_b2 ∧ ¬Tbet_b1)
IL18 = false
Tbet_b1 = (STAT1_b1 ∧ Tbet_b1 ∧ ¬STAT1_b2) ∨ (Tbet_b1 ∧ Tbet_b2 ∧ ¬STAT1_b1) ∨ (STAT1_b1 ∧ STAT1_b2 ∧ Tbet_b1 ∧ Tbet_b2) ∨ (STAT1_b1 ∧ STAT1_b2 ∧ ¬GATA3 ∧ ¬Tbet_b1) ∨ (STAT1_b1 ∧ ¬GATA3 ∧ ¬STAT1_b2 ∧ ¬Tbet_b1) ∨ (Tbet_b1 ∧ ¬GATA3 ∧ ¬STAT1_b1 ∧ ¬Tbet_b2) ∨ (STAT1_b1 ∧ STAT1_b2 ∧ Tbet_b1 ∧ ¬GATA3 ∧ ¬Tbet_b2)
STAT1_b2 = IFNgR_b1 ∧ IFNgR_b2 ∧ STAT1_b1
IL4R = IL4 ∧ ¬SOCS1
GATA3 = STAT6 ∧ ¬Tbet_b1
IFNgR_b2 = IFNgR_b1 ∧ IFNg_b1 ∧ IFNg_b2 ∧ ¬SOCS1
IL12 = false
IFNgR_b1 = IFNg_b1 ∨ (IFNgR_b1 ∧ IFNgR_b2 ∧ ¬IFNg_b1)
STAT4 = IL12R ∧ ¬GATA3
STAT1_b1 = IFNgR_b1 ∨ (STAT1_b1 ∧ STAT1_b2 ∧ ¬IFNgR_b1) ∨ (IFNbR ∧ ¬IFNgR_b1 ∧ ¬STAT1_b1) ∨ (IFNbR ∧ STAT1_b1 ∧ ¬IFNgR_b1 ∧ ¬STAT1_b2)
IL18R = IL18 ∧ ¬STAT6
IFNg_b2 = (IFNg_b1 ∧ Tbet_b1 ∧ Tbet_b2) ∨ (IFNg_b1 ∧ IRAK ∧ STAT4 ∧ ¬Tbet_b1) ∨ (IFNg_b1 ∧ IRAK ∧ STAT4 ∧ Tbet_b1 ∧ ¬Tbet_b2)
stable = b.stable_states
tabulate(stable, headers='keys', tablefmt="html")
| Tbet_b1 | GATA3 | IL4 | IFNg_b1 | IFNg_b2 | IFNbR | Tbet_b2 | IRAK | IFNb | STAT1_b2 | IFNgR_b2 | IL12 | IL4R | IL12R | SOCS1 | IFNgR_b1 | STAT4 | IL18 | STAT6 | STAT1_b1 | IL18R |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| False | True | True | False | False | False | False | False | False | False | False | False | True | False | False | False | False | False | True | False | False |
| False | False | False | False | False | False | False | False | False | False | False | False | False | False | False | False | False | False | False | False | False |
| True | False | False | True | False | False | False | False | False | False | False | False | False | False | True | True | False | False | False | True | False |
| True | False | False | True | True | False | True | False | False | False | False | False | False | False | True | True | False | False | False | True | False |
